High-Tech Software House
(5.0)
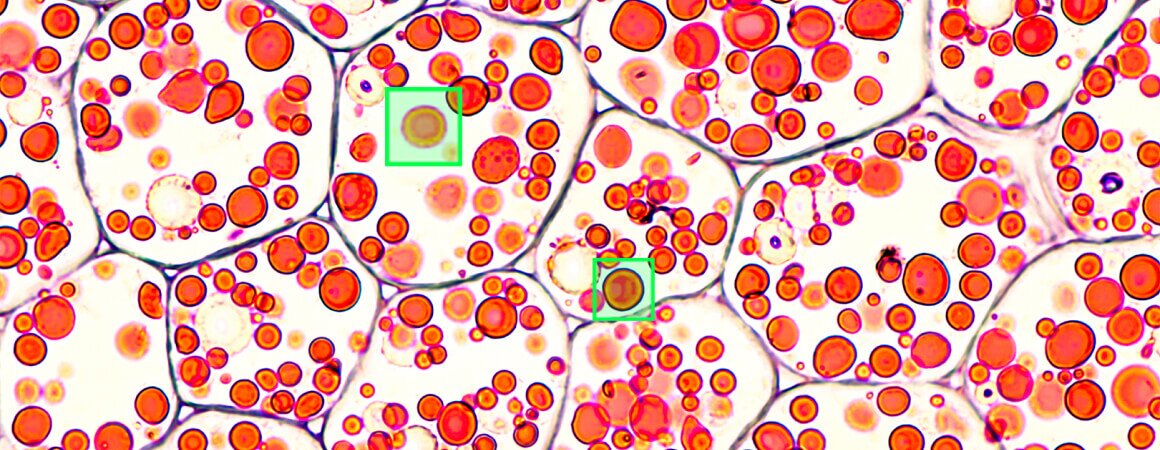
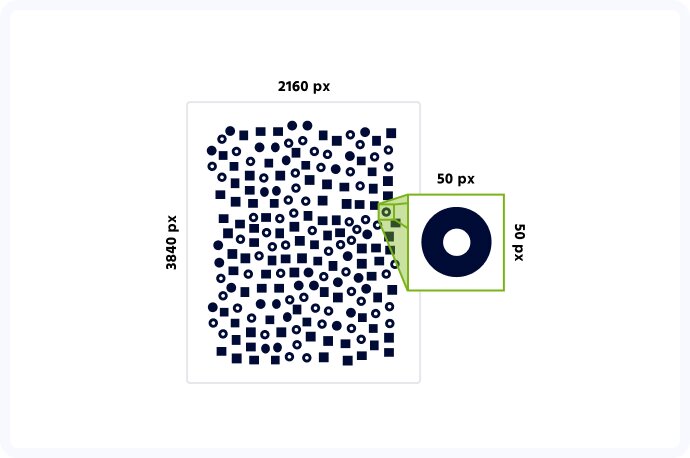
Computer vision is an incredible tool that can prove incredibly useful in many areas of everyday and professional life. Be it facial recognition and Instagram filters that change you into a unicorn, advanced intralogistics and warehouse automation or, you guessed it, computer vision in biotechnology and medicine. When we embarked on our cell detection project, our main goal was to create software that can quickly and accurately detect multiple cells from a high-resolution image, taken using a custom-built microscope. With the size of photographs measuring up to 4k pixels in each dimension, and the cells themselves being as small as 50 pixels each, you can probably imagine, it wasn’t an easy task. And yet, we did it!
When you stop to think about it, you can see how many incredible uses computer vision can have in healthcare and medicine. With advanced techniques like Mask-R Convolutional Neural Networks (Mask R-CNN), detecting brain tumor cells becomes much easier and quicker, canceling out the human error and aiding in the diagnosis process. Computer vision was even used during the current Covid-19 pandemic. Darwin AI located in Canada created a prototype application with the name of COVID-Net, which has shown results with around 92,4% accuracy in covid diagnosis. Cell detection aided by computer vision can be incredibly useful in healthcare research. It allows for accurate monitoring of cell colonies, which is crucial in determining the success rate of various substances, designed to kill or multiply those cells. Imagine a bacteria colony in a petri dish, that’s the main subject of new disinfectant research. With computer vision cell detection, we can quickly and accurately count the number of cells present before, and after applying the disinfectant, therefore measuring its effectiveness.
Our first goal in this project was to get the perfect cell count for a given sample. With this crucial aspect perfected, we could move forward to the next step of the development of effective computer vision-based cell detection. We wanted to localize each cell and perform segmentation to compute the percentage of biomass visible concerning this particular sample. Not only the number of cells is important in medicinal research, but also their shape and location. Those are crucial information that can save lives, and it’s computer vision that can help us make it happen.
The first major challenge we encountered was the incredible variety of the images taken. Some samples contained as little as 70 cells, while the others counted up to 1500! Naturally, accurate cell detection is the basis of this project, so it wasn’t an issue in itself but was a crucial element that we needed to take into account while designing an algorithm.
Other than the sheer magnitude of the cells visible, we had to take into account other elements that were unavoidably visible in the images. Things like stray bacteria, and of course, the calibration lines. In this project we used three different types of cells as our test material. Cells used in our tests were varying in color, size, and shape. That’s why we mentioned that in the practical use of computer vision in biotechnology and healthcare, it’s not just about the number of cells, but also about being able to specifically identify them.
Cells we used had different levels of illumination, sometimes based on their position - the ones in the center were slightly lighter (or darker in some cases) than the ones in the image corners. The focus was also the sharpest in the center, while you could see blurry edges when moving away from the middle. Some cells were in the background, which made accurate cell detection quite tricky. The last thing that we needed to consider was the relative position of the cells. In most of the cases, cells were neatly separated, but those cells have the tendency to cluster together, so there were no clear-cut boundaries between them. Simple background separation wasn't going to cut it.
Sometimes, the simplest solutions are the best. There’s often no need to overcomplicate things, and in complex biotech projects based on computer vision, time is often a crucial asset. With that in mind, before bringing out the big guns, we tried to use a "classical" computer vision approach. We constructed a pipeline that applied different color transformations, filters in HSV colorspace, and some more advanced morphological operations. With this simple approach, we managed to achieve decent results: ~75% accuracy for our validation dataset. However, in the later phases of the project, we had to write specialized code for each case that we found. As we mentioned, time is often of the essence, and writing a specialized code every time we need to use our software isn't going to be sustainable. Every change in lightning or focus significantly reduced our performance metrics. Our initial method was fast and didn't require any training steps, but finally wasn't robust enough and didn't generalize well.
The next approach that we tried was to check the segmentation architecture. Since it was built specifically for biomedical image segmentation, it seemed like a perfect candidate for our computer vision project. However, significant image size is a significant problem when applying deep learning algorithms. We needed a lot of GPU memory just to load the images during the training. Resizing them meant that we ended up with cell sizes measuring in single pixels, which wouldn’t allow us for accurate detection of shape and size. To alleviate this, each image was split into several smaller tiles. This way, we were able to keep memory requirements within a reasonable range and didn't lose any details from the original image.
We achieved significant improvements in comparison to our classical baseline. But as is often the case in the development process, we ran into a few other problems. At this point, our biggest challenge was to correctly separate individual cells when they were clustered together. Our neural network sometimes just didn't respect the boundaries between single cells, even when we applied weight maps in addition to simple binary output. And we all know that not respecting one's boundaries and personal space isn't a good thing. Even if it applies only to test cells. We had to think of a solution, to provide personal comfort to the cells taking part in our experiment, and effectiveness and accuracy for our computer vision project. For that, we needed to get as close to perfect segmentation as possible.
After much trial and error, we decided to create a hybrid approach, where we first detected the cells and then ran segmentation on each separate cell cut out from the original image. The detection step was done using a neuron model created specifically for detection and classification of organisms. Even though we got great results in most of the cases, we didn't have to worry to obtain perfect bounding boxes, because the segmentation step should compute the exact shape of each cell. Thanks to the reduction of the segmentation problem to single cells,the segmentation algorithm had no more issues with detecting the correct boundaries. Computer vision is about computer learning after all.
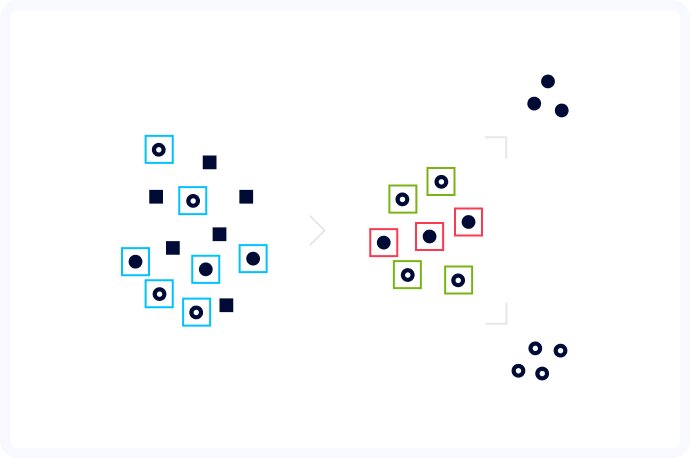
With this project, we were not only able to create powerful software with the potential to introduce computer vision into healthcare, but also gathered a lot of crucial experience for our team. We learned that segmentation algorithms don't work well when presented with images with a large number of instances so it's better to use them as a part of a pipeline with a detection algorithm as a first step. Arguably the most important aspect we discovered, is that when training a cell detection algorithm, the quality of the provided data is of the highest importance. It's better to have a smaller data set with high quality than a large DS with many errors. Hard sample mining can also be used to further improve overall detection quality. And finally, this kind of ML pipeline doesn't extrapolate well. When images with different properties (brightness/sharpness/noise) than the ones used for the training are supplied to the algorithm, the results are just plain bad.
Working on this project was a true delight for our team. Challenging, yet with a noble cause behind it. That’s what drives us forward. We hope to introduce more computer vision-based solutions into the constantly developing biotechnology field, and help with further medicinal research and healthcare uses of cell detection and computer vision overall.
If you’re further interested in the topic, make sure to use our contact form to send us a message! We love sharing our knowledge and experiences, as well as learning new things. So if you worked on a similar project, tell us about it! Sharing is caring after all, and caring makes the world a better place.
See our tech insights
Like our satisfied customers from many industries